Today I and my collaborators have a new paper published in PNAS! Justin Tackney, the lead author of the paper, was kind enough to write up a summary of the findings to publish here on Violent Metaphors. Here is his take:
The genetic characterization of the ancestral population in Beringia for the initial peopling event into North and South America has been limited by the lack of human remains buried at the right time and place. We report on two mitochondrial genomes (mitogenomes) from the Upward Sun River site in Alaska dated to about 11,500 years ago. This site is located in what was then eastern Beringia, and was occupied only a few thousand years after the main migration south, through an inland route, along the pacific coast, or both, about 15,000 years ago. The populations living at Upward Sun River at that time might represent a residual group that subsequently left or disappeared from the region today.
With the permission and support of the Healy Lake Tribal Council and Tanana Chiefs Conference, we obtained DNA from the remains of two individuals buried at the site. One was an infant just a few weeks old. The other was a late pre-term fetus.
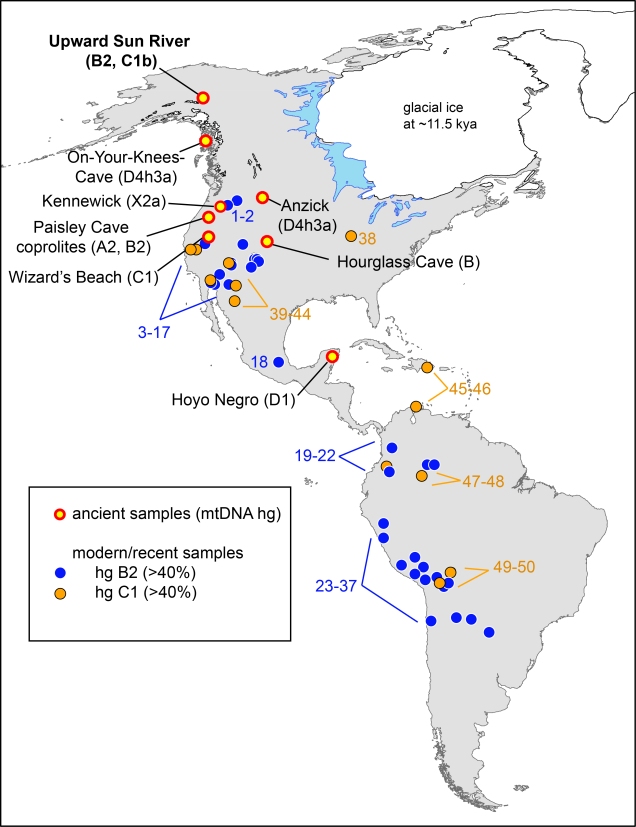
discussed. The locations of the Upward Sun River site, as well as the seven previously reported archaeological sites dated at >8,000 y B.P. with successfully genotyped human mitochondrial DNA lineages, are listed on the map (with reported haplotypes). Reported populations of ≥ 20 individuals with ≥ 40% C1 (yellow) or B2 (blue)
are shown.
While the two remains were initially believed by archaeologists to be maternally related, the infant and the pre-term fetus carried two different mitochondrial ‘haplotypes’, or mitochondrial DNA variants. This is significant because mitochondrial DNA is only maternally inherited, so it means that they did not share a mother. Both haplotypes, C1b and B2, are rare or absent in Northern North America today (although Jennifer, Dennis, and I did find B2 present in a ~1000 YBP population from the Brooks River area of the Alaska Peninsula in a previous paper*, suggesting that it was once more common in Alaska) and both were determined to be phylogenetically within or near the roots of their respective clades.
That these haplotypes were so far north at this very early date supports the Beringian Standstill Hypothesis (the idea that people lived in Beringia for at least several thousand years before moving southward into the Americas) by providing evidence for the ancestral root variants in the initial source population and by showing large genetic variation even within one residential complex. There was a third cremated 3-year old child found above the burial, dated to about the same time as the two human remains analyzed, but the cremation seems to have prevented the preservation of any DNA.
We were able to use the two precisely dated remains to inform the rate of the molecular clock within these two lineages and re-date the time to most recent common ancestor for all known Native American variants within these haplotypes. Our dates were on the younger side of recent estimates for these two clades (C1 and B2), but were close to previous dates calculated with a molecular clock also estimated with ancient DNA. We estimated C1b to coalesce at 12,854 (11,853-14,079) years before present and B2 to coalesce at 12,024 (11,500-14,085) years before present. These younger coalescence dates might be used to support a much shorter period of isolation in Beringia, as suggested by Raghavan and colleagues** earlier this year.
Our two mitogenomes are only the 8th and 9th mitochondrial lineages determined from human remains in the Americas dating to greater than 8,000 years ago and we are one of only 3 sites where the whole mitochondrial genome was sequenced completely. The two individuals at Upward Sun River are the farthest north human remains thus far discovered.
We performed hybridization capture for mitochondrial DNA molecules and then sequenced the captured ancient DNA on an Ion Proton system; we are one of the first groups in our field to use Ion technology for this purpose.
Do you have any questions for the research team? Leave them in the comments below, and either Jennifer or Justin will answer them.
References
*Raff JA, Tackney JT, O’Rourke DH. 2010. South from Alaska: A pilot aDNA study of genetic history on the Alaska Peninsula and the Eastern Aleutians. Human Biology 42 (5-6): 677-693.
**Raghavan et al. 2015. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science DOI: 10.1126/science.aab3884
Reblogged this on realitygenia and commented:
Thank you, your article was very interesting and insightful! I read and shared it with my network.
Your graphic shows B2 in the Paisley Caves coprolites (ca, 14,300 cal yr BP) but this is much older than the coalescence age you present here (even using the early end of the range of variation of ca. 14,100). Either your estimate requires tweaking or, perhaps, there’s something wrong with the Paisley Caves DNA (as Poinar et al. and I and Goldberg et al. have suggested).
Hi Stuart,
I don’t really want to wade into the Paisley cave authenticity argument, as I don’t have a horse in the race. I will admit, however, that you are correct that the coalescent dates we got using BEAST and the single dated USR2 sequence are younger than that reported for the coprolites. Some comments I feel appropriate/secure offering here:
I am open to the idea that our coalescent dates for the two clades might be a bit younger than reasonable, and we would certainly get more clarity with further whole mitogenomes within the B2 clade. Right now the BEAST analysis is just based on one dated node (USR2). I will remind you that our 95% HPD interval was 11,500–13,085.
Second, the most parsimonious tree has USR2 within the root of B2, particularly because of the ancestral allele at position 3547. If alternatively this represents an authentic back mutation from the derived G to A, this would allow a bit longer time for the coalescence of the B2 clade, and USR2 would be one mutational event away from the root. I must stress, however, that our results for C1b and B2, given the age of the two individuals, do point to more recent expansion times than that typically calculated by Maximum Likelihood analyses of modern sequences. The aDNA contribution here is key.
Third, I’ve always been very confused on what SNPs were actually typed in the original Gilbert et al. 2008 paper. I believe for the B2 specimens they first typed the 9bp deletion and then they typed T4977C and C11177T. That’s all. Where the Paisley cave specimens fall in the phylogenetic tree around the B2 root is unknown. Additionally (more importantly?) the B2 results were not replicated in the follow-up paper by Jenkins et al. 2012. We chose to keep the A and B2 typing from the 2008 paper for our Figure 1.
Thanks,
Justin
Justin,
Thanks for your rapid and informative response. For those who haven’t read my more recent comments about Paisley Caves in Human Biology (or at Academia.edu), I will reiterate two very odd aspects of the claimed human mtDNA (leaving aside the co-occurrence of canid DNA in the same samples). First, throughout the stratigraphic sequence which extends from claimed pre-Clovis until the late Holocene, only mtDNA haplogroups B and A are present, while C and D are absent. This is in sharp contrast to the relative frequencies found in both recent native peoples of the Great Basin and in ancient samples from the region. Second, many of the specimens with A markers also have the 9 bp deletion that characterizes B. This puts them in clade A2d1, found today only in a few Mexicans and Central Americans and, to my knowledge, not found in any recent or ancient native North Americans.
Stuart
Thank you, Stuart Fiedel, for your past writings on Native American writings and adding your voice to the feedback and debate here. I’m just a modest “Internet Journalist” who keeps my geneticist friend Simon Southerton apprised of goings-on in the area of advances of our understanding of how humans populated this hemisphere (Simon and I both grew up under the umbrella of Mormon Church claims and had to shed a lot of the nonsense we were taught as impressionable youngsters; he made a contribution to the on-line Encyclopedia of Global Human Migration, and I was privileged to have him share his work and thoughts with me as well as offering my feedback). I’m grateful you haven’t ducked the controversy on the “Monte Verde dates” as well (no prizes for guessing where I lean on that one).
Okay, back to my cubby hole.
Ugh… Need to proofread better because of lack of editing feature here. Should read: “Native American origins,” of course.
Randy,
Thanks for your supportive remarks.
SF
Jennifer,
You and your collaborators’ article received a nice write up in the local Salt Lake Tribune.
http://www.sltrib.com/home/3095008-155/utah-genetics-research-links-ancient-babies
I’ve listened to enough scientist friends over the years to understand their dismay when they’re “misrepresented or misquoted” in mainstream media coverage, but Ms. Moulton’s reporting looks spot-on this time. And instead of “pandering to the diffusionists out there,” she does reference legitimate debate on the subject of “The Berengia Standstill” and how long that period involved (no opinions here, honest, even if I have decided “Clovis First” tendencies).
Regards,
rw
Very enlightening, well written article. Thanks